Viral leukemia, monitor therapeutic response
Press release (05/09/2017)
Novel tool to monitor therapeutic response of patients with viral leukemia
Brussels, 5 September 2017 – A few months after the discovery of a novel mechanism by which viruses cause leukemia (Nature Communications, 23rd May 2017), the research team led by Anne Van den Broeke (Institut Jules Bordet - University of Brussels and GIGAUniversity of Liège) publishes a new study that addresses a clinical question. In this pilot study, published 5 September in the journal Leukemia, the researchers apply an optimized molecular method to monitor disease progression in patients. They show that the new method enables a better evaluation of the therapeutic response for this particularly aggressive type of leukemia.
From basic research to translational studies
To address basic questions in cancer development, Anne Van den Broeke’s team is studying a virusinduced leukemia caused by the Human T-cell leukemia virus-1 (HTLV-1) in combination with the corresponding animal model of blood cancer associated with infection by the closely-related Bovine Leukemia virus (BLV, a virus infecting cattle and sheep). In the fundamental study published in May 2017, the team developed several novel methods based on next generation sequencing (NGS) technologies which resulted in the discovery of a novel mechanism by which oncogenic viruses contribute to cancer development. In analyzing the data, they realized that one of the methods they had been improving during this study (it maps the exact position of the virus within the genome (genetic material) of the host) could be of interest for monitoring patients under therapy. The team then put all their efforts in optimizing the method.
A method with clinical applications
Patients with unfavorable viral leukemia subtypes have a poor prognosis. Clinical remission and response to treatment are currently defined on the basis of consensus criteria that have not been revised over the eight years since they were first described. Given the high rates of rapid relapse after achieving remission, there was an unmet need for improved molecular tools to better estimate response to treatment. At diagnosis, each patient has a “viral ID card”, which corresponds to a precise location of the virus in the genome of their blood cells. The NGS method quantitatively detects this position. In patients under treatment, its decrease reflects an good therapeutic response while an increase reveals recurrence of the disease. The pilot study published in Leukemia reports the follow-up of patients using the optimized approach developed by Van den Broeke’s team. The authors found that the method can reveal patients who are refractory to first-line therapy. In addition, it enables the detection of early relapse that escapes detection by conventional methods and outperforms any other molecular method examined thus far. Importantly, besides improving its sensitivity, the researchers dramatically reduced the cost of the technique, facilitating its implementation for clinical use. The study, carried out in collaboration with scientists of the Necker University hospital in Paris, is a nice example of fundamental research that leads to a clinical application. Joint efforts of the HTLV-1 community of clinicians will now be needed to define optimal conditions for clinical utilization and standardized care to the benefit of the patient.
- Reference of the study
Maria Artesi, Ambroise Marçais, Keith Durkin, Nicolas Rosewick, Vincent Hahaut, Philippe Suarez, Amélie Trinquand, Ludovic Lhermitte, Véronique Avettand Fenoel, Michel Georges, Olivier Hermine & Anne Van den Broeke Monitoring molecular response in Adult T-cell Leukemia/Lymphoma by high throughput sequencing analysis of HTLV-1 clonality, Leukemia, 2017, doi:10.1038/leu.2017.260
- Senior investigators
Dr Anne Van den Broeke (Institut Jules Bordet and GIGA) and Prof Michel Georges (GIGA).
Laboratory of Experimental Hematology, Institut Jules Bordet, Université Libre de Bruxelles (ULB), Boulevard de Waterloo 121, 1000 Brussels, Belgium.
Unit of Animal Genomics, GIGA-R, Université de Liège, Avenue de l’Hopital 11, B34, 4000 Liège, Belgium.
- Research funding
This work was supported by les Amis de l’Institut Bordet, the Fonds de la Recherche Scientifique (FRS), Télévie, the International Brachet Stiftung (IBS) and the Fondation Lambeau Marteaux.
- International collaborations
Hôpital universitaire Necker, Paris, France – Dr Olivier Hermine and Dr Ambroise Marçais
- Enclosed :
Patients’ follow-up illustration
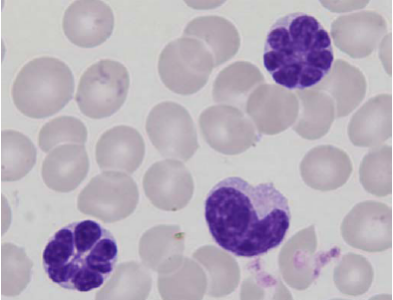
Picture : Typical “flower cells” in the blood of patients with viral leukemia
- For more information:
Dr Anne Van den Broeke: anne.vandenbroeke@bordet.be
Prof Michel Georges: michel.georges@ulg.ac.be
Press contacts
- Institut Jules Bordet
Ariane van de Werve
Tel: +32 2 541 31 39
GSM : +32.48617 33 26
E-mail : ariane.vandewerve@bordet.be
www.bordet.be
- University of Liège
Didier Moreau
Tel +32 4 366 52 17
GSM : +32 494 572 530
E-mail : dmoreau@ulg.ac.be
To download
- Institut_Jules_Bordet_Leucemie_nouvel_outil_prédictif_reponse_traitement.pdf136.84 KB
- Institut_Jules_Bordet_Novel_tool_monitor_therapeutic_response_patients_viral_leukemia.pdf145.53 KB
- Jules_Bordet_Instituut_nieuwe_tool_voorspellen_behandelingsrespons_virale_leukemietypes.pdf147.05 KB
- Flower_cells_ATL.jpg106.31 KB
- Leucemie_Illustration_suivi_patients.pdf260.16 KB
- Leukemia_Patients_follow-up_illustration.pdf338.99 KB